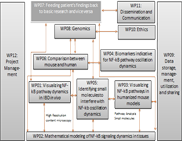
WP04 - Biomarkers indicative for NF-kappa-B pathway oscillation dynamics
Objectives
The key objective of this work package is to identify inflammatory biomarkers that together can be used to define and predict disease aetiology. This WP will build on knowledge developed on model systems in WP01 and WP03 and on the modelling of these systems in WP02. A combination or cell and tissue culture models together with detailed gene expression profiling will be used and dynamic properties of the signalling system inferred using modelling and data simulations.
- Define robust reporters of NF-kB network activation in cell and tissue samples.
- Use gene expression profiling to correlate NF-kB signalling to the inflammatory response.
- Use data from objective 1 & 2 and modelling from WP02 to identify biomarkers for IBD.
Workpackage Description
The concept of biomarkers is encapsulated within the definition of the National Institute of Health (NIH, US) as a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. IBD has a complex and variable patient specific pathogenesis and largely because of this existing biomarker have modest success in predicting clinical outcomes. To date, biomarkers for IBD have focussed on three main areas: (i) serum concentration of pro-inflammatory (TNF-alpha, IL6) and anti-inflammatory (IL10) cytokines; (ii) serum levels of C-reactive protein (CRP), a biomarker for general inflammatory status and (iii) biomolecules from microorganism in the gut biota.
Our objective is to identify and measure components of the NF-kB pathway that correlate with pathway activation (induction) as defined by the oscillation dynamics of the signalling process. We will explore the expression and behaviour of key signalling components (e.g. receptor expression, signal transduction proteins and reprogramming gene expression) and use modelling to identify and validate candidate biomarkers. This will be done in tissue culture and animal models (WP01), and tissue samples from patients (WP06) analysed in tissue culture and human-mouse xenografts (WP03). Samples from relevant models will be analysed by gene expression profiling both as cell populations and as single cells to reveal potential markers of persistent inflammation. Modelling of the signalling network and alterations in NF-kB target gene expression will then be analysed and modelled (WP02) in order to identify robust changes in the minimal set of components that best define the inflammatory status.
Task1: NF-kB network architecture in cell and tissue culture models
Changes in the intra-cellular location and concentration of NF-kB components provide excellent prospective biomarkers for persistent inflammation when used in combination with other inflammatory reporters (Fig. 1). Notably, the I?B proteins are degraded when NF-kB is activated so alterations in IB expression and dependent location of the NF-kB transcription factor proteins map precisely to activation of NF-kB target genes. Experimental data derived from WP01 & 03 and modelling from WP02 will confirm the architecture of the NF-kB network in a range of cell and tissue culture models. Potential biomarkers will be defined based on the analysis of the NF-kB network architecture using time-lapse microscopy of cells expressing fluorescent NF-kB proteins, immuno-cytochemistry to label components using antibodies (this will allow analysis of archival tissues) and flow cytometry. Our analysis will focus on 3 signalling components which are thought to modulate context-dependent inflammatory signalling; (i) the cell surface expression of receptors for established biomarkers TNF-alpha and IL-6 and other receptors that induced signalling through the NF-kB; (ii) dynamic properties of the signalling network proteins and (iii) the downstream changes in expression of target genes. Candidate biomarkers will be assessed in appropriate tissues and validated using patient samples during treatment programmes using cell biology techniques and animal studies using clinical samples.
Task 2: Changes in gene expression linking NF-kB activation to the inflammatory response
The NF-kB signalling network is a major regulator of inflammation with >300 known target genes, which have variable responses to different inflammatory inputs. In tissues, different cell types that contribute to persistent or chronic inflammatory diseases, such as IBD, secrete different combinations of pro-inflammatory cytokines and chemokines. Using cells treated in defined ways to activate NF-kB signalling through the canonical and alternative pathways and appropriate cells and tissue from animal models and patients, we will perform a time dependent analysis of target gene expression (reprogramming) using ChIP Seq to monitor dynamic changes at target promoters and HiSeq RNA sequencing to monitor changes in target gene transcription. For the latter approach, GRO-seq (metabolic labelling after a defined stimulus in order to detect newly synthesised transcripts) will be performed. The results will help to explain how the inflammatory input driven by activation of the NF-kappa-B and other inflammation-inducing pathways and the physiological output alterations in target gene expression - are linked. To define key links, gene expression profiling will be performed in patient samples and specific cell types isolated from these samples. The latter analysis can be performed on cell populations or individual cells, in order to provide a strong indication of stochastic events that contribute to the inflammation-dependent changes in gene expression. Data derived from these experiments will be modelled to identify likely control point in the signalling network and the most significant changes in expression as candidate biomarkers.
Task 3: Modelling the inflammatory response to identify biomarkers for IBD
As a key part of this analysis, mathematical models from WP02 will be used to predict the dynamic behaviour of NF-kB system components in fixed (e.g. cell culture, tissue or archival) samples. In patient cohorts, we will measure potential biomarkers in available tissues samples (e.g. gut, blood, stools) to identify the status of activation of the NF-kB-dependent inflammatory response. In addition, NF-kB pathway components will be measured in isolated and purified cell populations of the gut tissue and blood to define the NF-kB pathway dynamics in different cell type and tissue compartments during active disease, remission and controls. A major goal for development of predictive diagnostics biomarkers is to better predict increase in disease activity, complications, and co-morbidities. The deliverables of WP04 are therefore to measure the concentrations of NF-kB pathway components in tissue correlate these concentrations to the dynamics of the pathway and identify the best correlating components for further development of new diagnostic biomarkers. Candidate biomarkers will be validated in appropriate tissue samples and correlated with NF-kB activation and the severity of the inflammatory response.
Task 4: Long-term prospects
In IBD, treatment programmes are often complex because the contribution of genetic and environmental factors to disease progression is difficult to predict. As pro-inflammatory cytokines such as TNF and IL6 are major drivers of IBD these proteins are key targets for therapeutic intervention. Humanised antibodies to TNF (e.g. infliximab) are now in common use across EU and often have excellent outcome in combination with other drugs (e.g. steroids). But drugs such as infliximab are expensive (estimated cost of infliximab is ~$20,000/patient/year) and the benefits of treatment can be limited by the expression of neutralising antibodies which develop in the patient during prolonged period of treatment. By looking at the behaviour of NF-kB signalling in tissue culture and animal model systems we will develop a detailed understanding of the disease progression and begin to use this knowledge to optimise therapy protocols. At present, the most effective drugs for IDB therapy target the inflammatory input i.e. through TNF and TLR receptors and features of tissue architecture related to migration of the inflammatory cells. Pharmaceutical companies such as GSK and AZ have developed other potential inhibitors of the NF-kB signalling network (e.g. to IKK and GSK3-beta) and we will test such drugs to evaluate the most effective treatment combinations. The knowledge generated using these systems to study the inflammatory process in IBD models has the potential to deliver novel insights into the best treatment strategies for IBD and eventually significantly improve the quality of life of affected individuals across EU and the world.
Over the corse of the project, by combining biological measurement with mathematical modelling and use of sophisticated databases, we will define therapeutic intervention strategies to modulate NF-kB signalling in a logical and targeted way.